RNA-Seq: The Cells Librarian
A DIY Guide To Analyzing RNA-Seq Data For Differential Expression Analysis
An ask: If you liked this piece, I’d be grateful if you’d consider tapping the “heart” 💙 in the header above. It helps me understand which pieces you like most and supports this newsletter’s growth. Thank you!
🧫 RNA-Seq: The Cell’s Librarian
In the past, I worked with a professional cyclist, serving as their personal physiologist and data analyst. Around that time, this athlete participated in a study comparing genetic markers for muscle fiber composition and actual measures of muscle fiber composition obtained via muscle biopsy.
Unsurprisingly, this athlete's muscle biopsy showed a high percentage of type-1, slow-twitch muscle fibers. However, we were surprised that this athlete had variants of the ACTN3, MYH, and ANGPTL4 genes associated with a higher-than-average fast-twitch muscle fiber composition. Despite this athlete's genes indicating a predisposition for a sprinter's phenotype, the reality was that they were an elite endurance with a dominance of slow-twitch fibers.
The apparent mismatch between this athlete's genetic predisposition and their actual phenotype stems from the dynamic nature of gene expression and the impact of environmental factors. While genetic tests offer insights into potential traits encoded in an individual's DNA, they often overlook the intricate interplay between genes and environmental stimuli. In the case of our endurance athlete, their extensive endurance training had triggered alterations in gene expression, favoring the development of slow-twitch muscle fibers—ideal for prolonged activities.
This experience prompted me to question the value of genotyping in elite athletes, which was gaining popularity at the time. Rather than myopically focusing on an athlete's genetic predispositions, I wanted to understand how gene expression changes in response to environmental factors, pharmaceuticals, and specific exercise interventions. This led me to RNA-Seq, a powerful tool for exploring gene expression. In a sense, you can think of RNA-Seq as a cellular librarian.
Imagine your DNA is a vast library containing countless books (genes), and each book contains instructions to create different elements in your body, like proteins. When a cell needs to make a specific protein, it checks out the relevant book and makes a copy (mRNA) through a process called transcription. RNA-Seq is a technology that enables us to quantify these mRNA copies. It's like a librarian who systematically catalogs the number of times each book is checked out and copied, providing insights into the goings on within the library at that moment.
In this article, I'll teach you what RNA-Seq is, how it works, and how you can analyze RNA-Seq data with basic programming skills. Let's dive in.
🧫 How Does RNA-Seq Work?
RNA-seq is a cutting-edge technique used to analyze the expression of genes in a biological sample. It works by converting RNA molecules into complementary DNA (cDNA), sequencing this cDNA, and mapping the sequence data to a reference genome, or transcriptome, to quantify gene expression levels.
Performing RNA-Seq in humans involves initially acquiring a tissue or blood sample for subsequent RNA extraction. The RNA extraction process entails lysing cells to release RNA molecules from the sample; then, the liberated RNA is transcribed into DNA through reverse transcription, where an enzyme generates a complementary DNA strand based on the RNA template. Subsequently, the resulting DNA undergoes sequencing, often via high-throughput technologies such as next-generation sequencing, a topic extensively covered in a previous article titled, From Biomolecules to Bytes.
After sequencing, a digitized file is outputted in a standard format, such as FASTQ, and includes the sequence information, quality scores, and metadata for each read. Bioinformatic analyses are conducted from these files to map the sequences back to a reference genome or transcriptome, which refers to the complete set of all RNA molecules, including messenger RNA (mRNA), non-coding RNA, and other RNA species produced in one or a population of cells. By mapping our sequence to a reference transcriptome, we can count how many reads align to a given region and deduce the original amounts of RNA in the cell, providing valuable insights into gene expression levels.
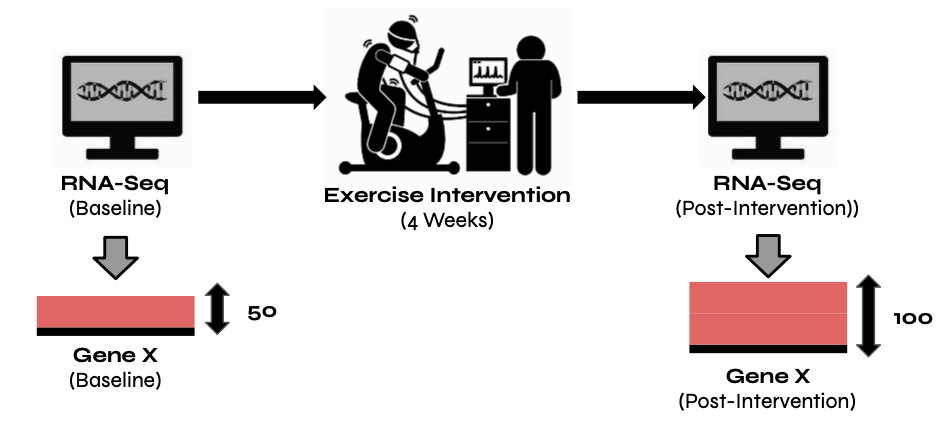
One of the biggest values of RNA-Seq, in my opinion, is that you can collect samples at different time points to see how gene expression changes in response to different environmental conditions (i.e., cold shock), therapeutics, or exercise interventions, as demonstrated in the image below.
In the image above, we see Gene X's expression doubling following an exercise intervention. Understanding whether a gene is up-regulated or down-regulated by a statistically significant degree is only the start when trying to understand RNA-Seq data. The next step is to try and understand what that data means from a biological perspective. For example, upon seeing Gene X’s expression level doubling in response to a specific exercise intervention we should ask ourselves what the known functions of Gene X are and whether its role in exercise has previously been identified. If Gene X's role in exercise is known, our analysis further validates previous findings. However, if it's not known, we may have found a new role for Gene X that needs to be explored further.
Additionally, in the real world, we're often looking at how thousands of genes are differentially expressed following an environmental stimulus, exposure to a drug, or some other intervention. For example, let's take the image below, generated with the GEO2R analysis tool using data from a study titled, Amiloride, An Old Diuretic Drug, Is a Potential Therapeutic Agent for Multiple Myeloma.
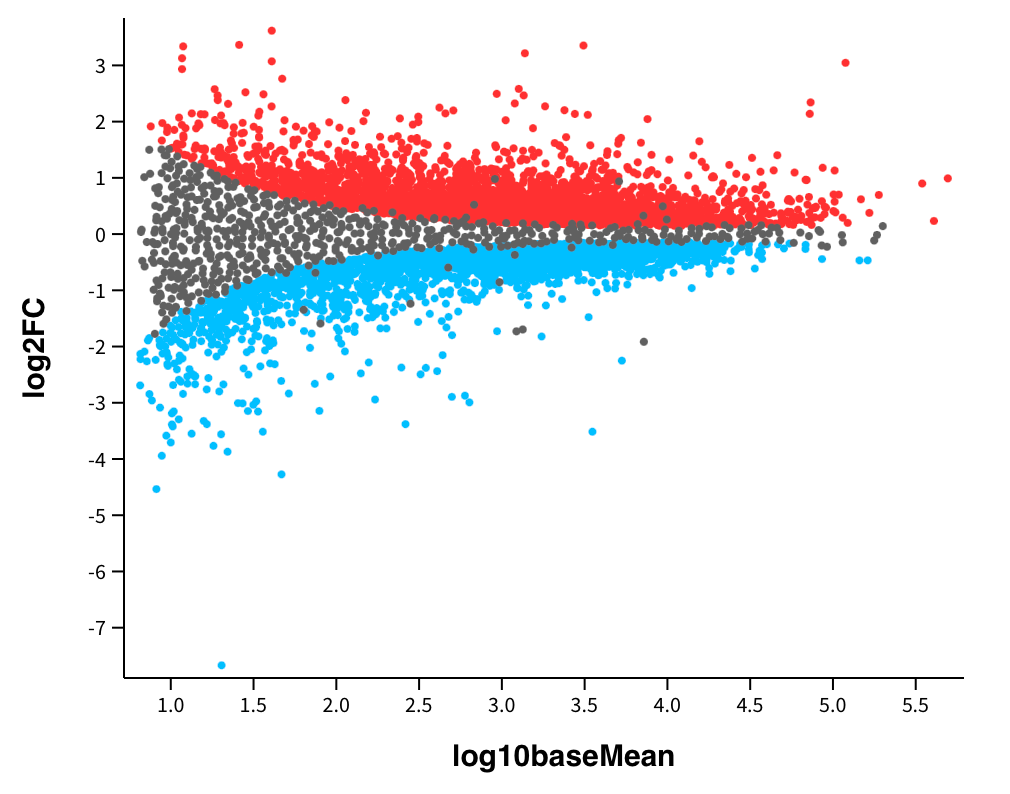
The chart above shows the fold change on the y-axis, which represents the magnitude of the change in gene expression between the control and treatment conditions. The fold change is calculated as the ratio of the expression level in one condition to the expression level in another, and it is used to assess the biological significance of gene expression changes. Each data point on the chart above represents a specific gene. Genes highlighted in blue are down-regulated in the treatment group compared to the control group, and genes highlighted in red were unregulated in the treatment group compared to the control group.
When assessing the effects of a specific intervention, we need to consider its effects on individual genes and how these varying effects interact with one another. In Amiloride's case, analyses show that it significantly reduces the expression of some cancer-promoting genes but increases the expression of others. For example, in certain situations, Amiloride can inhibit ion channels involved in cancer cell proliferation and migration, highlighting its anticancer potential. In other situations, Amiloride may promote cancer progression by influencing tumor microenvironments. Through RNA-Seq studies, we can better understand how specific interventions impact gene expression over time.
🧫 How Is RNA-Seq Used In Biotech ?
RNA-Seq is a powerful tool widely used in research and the biotech industry to study gene expression patterns and it provides a comprehensive snapshot of the transcriptome, offering insights into how genes are activated or silenced. In medicine, RNA-Seq can help researchers identify disease-associated genes, unravel disease mechanisms, and discover potential therapeutic targets. For example, RNA-Seq has been used to identify genes linked to cancer, neurodegenerative disorders, and infectious diseases, which may be prime drug targets.
Additionally, RNA-Seq can be used in exercise physiology to elucidate the molecular response of muscles to exercise, offering insights into training adaptations, fatigue mechanisms, and potential targets for enhancing athletic performance. Researchers may even use RNA-Seq to spot potential therapeutic targets for exercise-mimetic drugs by identifying how genes are differentially expressed in response to different types of exercise interventions.
Furthermore, RNA-Seq has uses in agriculture, where it contributes to crop improvement by deciphering gene expression in response to environmental factors, thus guiding the development of crops with enhanced yields, resistance to diseases, and adaptability to diverse conditions.
In summary, RNA-Seq is a versatile tool that unveils the dynamic language of gene expression, facilitating breakthroughs across diverse fields from medicine to agriculture and beyond.
🧫 A DIY Guide To RNA-Seq
So far, we’ve covered what RNA-Seq is, how it works, and how its used in research and the biotech industry. In this section, I will provide an end-to-end DIY guide to analyzing RNA-Seq data. Specifically, I’ll show you how to analyze RNA-Seq data obtained via next-generation sequencing to better understand how genes are differentially expressed in response to certain experimental conditions.
💻 Step 1: Quality Control
The first step in analyzing RNA-Seq data is to assess the quality of the raw sequencing data to identify potential issues that may impact downstream analyses, thus ensuring your results are biologically meaningful and reproducible. To perform quality control of raw sequencing data, you’ll need FASTQ files representing nucleotide sequences and their corresponding quality scores generated through next-generation sequencing. In the code sample below, I’ll show you how to use FastQC, a command-line tool for quality control analysis of high-throughput sequencing data that generates quality reports for each sample in your FASTQ file:
fastqc S1R1.fastq.gz S1R2.fastq.gz -o fastqc_output/The code above runs FastQC on two paired-end FASTQ files and stores the analysis results in the “fastqc_output/” directory. Let’s break it down step-by-step to see how it works:
The code begins with the fastqc command, which runs the FastQC command-line tool. FastQC performs a comprehensive analysis of the input FASTQ files and assesses various aspects of the data quality, including per-base sequence quality, per-sequence quality scores, GC content, sequence length distribution, and the presence of sequencing adapters.
Next, the two input files (S1R1.fastq.gz and S1R2.fastq.gz) for the fastqc command are specified. These files contain nucleotide sequences of RNA fragments, along with quality scores for each base. Importantly, you should customize the file names to match the actual names of your FASTQ files before using the block of code above.
Finally, -o fastqc_output/ specifies the output directory where FastQC will generate its results.
After running the FastQC command-line tool, you can view the generated HTML and text files containing quality metrics and visualizations stored in the fastqc_output/ directory. These reports will provide a detailed overview of the quality characteristics of the input sequencing data. If potential issues such as poor sequencing quality or adapted contamination are identified, you can perform additional preprocessing before proceeding with downstream analyses such as read trimming or filtering.
💻 Step 2: Trimming and Filtering
After performing quality control, the next data preprocessing step is to trim and filter raw sequencing data, which involves removing low-quality bases, adapter sequences, and other artifacts from the raw sequencing data. The primary purpose of this step is to enhance the quality of the data before downstream analyses, leading to more accurate and biologically meaningful results in the exploration of gene expression patterns and regulatory mechanisms. In the sample code below, I’ll show you how to use the trimmomatic command-line tool to trim and filter raw sequencing data:
trimmomatic PE S1R1.fastq.gz S1R2.fastq.gz \
S1R1_trimmed.fastq.gz S1R1_unpaired.fastq.gz \
S1R2_trimmed.fastq.gz S1R2_unpaired.fastq.gz \The code above uses the trimmomatic command line tool to take paired-end data from S1R1.fastq.gz and S1R2.fastq.gz, perform quality trimming, and output the trimmed and unpaired reads into separate files for both forward and reverse reads. Let’s break it down step-by-step to see how it works:
First, the trimmoatic command is used to run the trimmomatic tool, and then PE specifies that the input data consists of paired-end reads.
Next, S1R1.fastq.gz and S1R2.fastq.gz refer to the input files containing paired-end reads in FASTQ format.
Then S1R1_trimmed.fastq.gz contains the trimmed paired-end reads, and S1R1_unpaired.fastq.gz contains the unpaired reads generated by trimmomatic for the forward reads after trimming. Similarly, S1R2_trimmed.fastq.gz and S1R2_unpaired.fastq.gz are the output files for the reverse reads.
Generally, specific quality trimming parameters, adapter clipping, and other settings are included as well, based on the specific needs of an analysis. However, I left these absent since these parameters are optional and lack uniformity.
💻 Step 3: Aligning Reads To A Reference Transcriptome
The next step in analyzing RNA-Seq data is to map the sequence reads obtained via high-throughput sequencing to a reference transcriptome, which involves aligning short RNA-Seq reads to annotated transcripts, enabling the identification of the origin and abundance of each transcript in the sample. Aligning reads to a reference transcriptome facilitates downstream analyses, such as quantifying gene expression levels and identifying differentially expressed genes. Additionally, accurate alignment is crucial for understanding the transcriptional landscape and interpreting the biological significance of gene expression patterns in the studied biological conditions.
To perform this step of RNA-Seq analysis, you’ll need both trimmed FASTQ files from the previous step and an organism-specific reference transcriptome in FASTA format. In the code below, I’ll show you how to align your align reads to a reference transcriptome using the hisat2 command-line tool:
hisat2 -x reference_transcriptome.fasta -1 S1R1_trimmed.fastq.gz -2 S1R2_trimmed.fastq.gz -S sample1.samThe code above uses the hisat2 aligner to map paired-end RNA-seq reads to a reference transcriptome. Now, let’s see how it works:
First, the hisat2 command is used to run the hisat2 aligner, which is commonly used for aligning RNA-seq reads to a reference genome or transcriptome.
Next, the code -x reference_transcriptome.fasta specifies the reference transcriptome. Following that, -1 S1R1_trimmed.fastq.gz and -2 S1R2_trimmed.fastq.gz specify the input files containing paired-end reads. -1 and -2 refer to the forward and reverse read files, respectively.
Finally, -S sample1.sam specifies the output file to store the alignment results in SAM format, containing information about the alignment of reads to a reference.
Notably, the SAM output file generated for each sample in an analysis referenced above can be converted to a BAM format, as demonstrated in the code below, making it easier to sort:
samtools view -bS sample1.sam | samtools sort -o sample1_sorted.bam -First, the code above uses the samtools view -bS command to take an input SAM file (sample1.sam) and convert it into the BAM format. Then, the code uses the pipe operator | to redirect the output of the previous code as input to the next command, which uses samtools sort to sort the BAM output from the previous command and then store it in a new file named sample1_sorted.bam.
The sample1_sorted.bam file can then be indexed with the following code, which enables quick access to specific genomic regions, allowing for efficient data retrieval:
samtools index sample1_sorted.bam💻 Step 4: Counting Gene Expression
The next step in analyzing RNA-Seq data is quantifying the abundance of transcripts or genes within a biological sample. This process involves assigning the sequenced reads to specific genomic features, such as genes or transcripts, and counting the number of reads associated with each feature. The resulting count data provides a quantitative measure of gene expression levels, reflecting the activity of genes in a given experimental condition.
By comparing expression profiles between different samples or conditions, you can identify differentially expressed genes, uncover biological pathways, and gain insights into the molecular mechanisms underlying various biological processes. Additionally, counting gene expression is a crucial component of RNA-Seq analysis, serving as the basis for downstream statistical analyses and identifying key genes associated with specific physiological or pathological conditions.
In the sample code below, I’ll show you how to quantify the number of reads aligned to each gene to create count tables using a sorted and indexed BAM file and a gene annotation file as input:
featureCounts -a annotation.gtf -o counts.txt sample1_sorted.bamThe code above uses the feature counts tool to quantify the number of reads assigned to genomic features, such as genes, based on the alignment information in a sorted BAM file. After calling the featurecounts command, the code -a annotation.gtf specifies the annotation file stored in GTF format, which contains information about genes and their coordinates on the reference genome.
Next, the code -o counts.txt specifies the output file where the count results will be saved. The counts represent the number of reads assigned to each genomic feature, which is used to generate a count table. Finally, the code section sample1_sorted.bam refers to the input BAM file containing the sorted and aligned RNA-seq reads that are aligned to a reference transcriptome.
💻 Step 5: Differential Expression Analysis
Differential expression analysis in RNA-Seq is a critical step that helps researchers identify genes whose expression levels significantly change between different experimental conditions or sample groups, which can help researchers identify potential biomarkers, elucidate molecular responses to different environmental conditions, drugs, or exercise interventions, and guide future experimental investigations.
One of the most common tools for performing differential expression analysis is DESeq1, a commonly used R package that allows you to generate results tables from RNA-Seq data. Because of my limited familiarity with this tool, at the time of my writing this article, I will not go into meaningful depth as to how to best use it. Instead, I will refer you to a previous article I wrote titled A DIY Guide To Differential Expression Analysis, where I'll teach you what differential expression analysis is, when and why it's used, and how you can perform it yourself.
🧫 Limitations Of RNA-Seq Analysis
RNA-Seq is undoubtedly a revolutionary technology for studying gene expression in response to different environmental conditions, therapeutics, and interventions. However, it's not without its challenges. Before wrapping this article up, I wanted to acknowledge some limitations of RNA-Seq analysis so as not to paint it as a magic tool for answering any and all biological questions.
One of the biggest limitations of RNA-Seq is that batch effects introduced during sample processing, library preparation, or different sequencing runs can introduce noise, impacting the accuracy of differential expression analysis. These effects can be large enough to impact the biological interpretation of data, which is why quality control is of such high importance.
Another limitation of RNA-Seq is that it's very difficult to accurately quantify the expression level of genes with lower-than-average baseline expression, making it challenging to detect differential expression between varying treatment conditions for these genes. This is a major concern since there may be genes of high importance for understanding the molecular basis of disease whose expression levels simply cannot be accurately detected with RNA-Seq. Furthermore, more systemic issues, such as incomplete reference datasets, can limit the effectiveness of RNA-Seq analysis. For example, reference transcriptomes used to align reads may not be fully annotated, leading to gaps in coverage or the omission of novel transcripts, which can ultimately impact the results of analyses.
Despite these limitations, RNA-Seq remains a widely used and valuable tool for gene expression studies. Careful experimental design, appropriate quality control, and consideration of statistical methods can help mitigate some of these challenges and improve the reliability and interpretability of RNA-Seq data.