Predicting Clinical Dementia Ratings From fMRI Data
An End-To-End Case Study Using Machine Learning To Predict Clinical Dementia Rating Scores From Health Data
The most recent Decoding Biology newsletters have been about deconstructing machine learning systems into their constituent parts to examine how they work. Today’s newsletter, on the other hand, will be about the big picture.
After loading the Oasis-1 dataset, containing cross-sectional MRI data for young, middle-aged, non-demented, and demented older adults, I’ll walk you through the processes of summarizing and preparing the data, selecting and evaluating algorithms, tuning hyperparameters, and finalizing a model to make predictions on new unseen data.
Below you’ll find a concept model showing how the various processes without our system interact:
However, I’d be remiss not to mention that the process of creating a machine learning system is not linear. Sometimes, you may loop between a subset of steps, skip steps, or backtrack to improve a model’s accuracy (as I do in this newsletter). Thus, you can consider the code below a first-pass through this machine learning project to establish a baseline model that can improve over time.
My recommendation to readers is to use a similar process, making a quick first pass through all of the steps above to get the process out of your head and into your coding environment, then cycle back through the steps attempting to improve your model’s performance and modifying your code as needed. If you’d like me to elaborate on specific steps in more detail, feel free to ask a question in the comment section as the bottom of this page.
🧬 Load, Summarize, and Clean The Data:
The Oasis-1 dataset contains cross-sectional MRI data for young, middle aged, non-demented and demented older adults. The block of code below demonstrates how to load, clean, and summarize the dataset:
Below you’ll find a list of attributes contained in the dataset after removing rows with missing data:
M/F: Male or female (0 = male, 1 = female)
Age: The subject’s age in years (note, the age range is 33-96 years, with the mean age being 72 years old)
Edu: The subject’s education level
SES: The subject’s socioeconomic status
MMSE: Mini mental state examination score
eTIV: Estimated total intracranial volume
nWBV: Normalized whole brain volume
ASF: Atlas scaling factor
The 8 factors listed above are the inputs variables that will be used to predict the outcome variable of interest, which is CDR in this dataset. CDR, is a clinical dementia rating score, which is scored from 0-3 as follows:
CDR of 0 = No dementia
CDR of 0.5 = Questionable dementia
CDR of 1 = Mild cognitive impairment
CDR of 2 = Moderate cognitive impairment
CDR of 3 = Severe cognitive impairment (note, there are no CDR=3 samples in the dataset, which is a limitation)
Since some machine learning algorithms, such as linear and logistic regression, suffer poor performance when the attributes in the dataset are highly correlated, it's a good idea to review all of the pairwise correlations in the dataset. You can easily do this with the Panda’s corr() function, as demonstrated below:
A handful of moderate to strong associations exist between attributes in the dataset. For example, nWBV is moderately associated with MMSE and strongly associated with age. This is often unavoidable when working with biomedical data. Thankfully, many of the attributes in the dataset are uncorrelated, so the existing correlations should have minimal impact on the model's results.
🧬 Spot-Check Algorithms:
Spot-checking is a way to quickly discover which algorithms perform well on your machine-learning problem before selecting one to commit to. Generally, I recommend that you spot-check five to ten different algorithms using the same evaluation method and evaluation metric to compare the model's performance.
In the code below, I use k-fold cross-validation as my evaluation method, with mean squared error (MSE) as my evaluation metric. Generally, accuracy and MSE are used as evaluation metrics for classification and regression problems, respectively. Technically, the problem I'm working on in this article is a multi-class classification problem. However, I'm treating it as a regression problem since I'm interested in predicting intermediate CDR values (ex, 2.4 versus 2 or 3 with no in-between). Hence, why I'm using MSE as my evaluation metric and spot-checking algorithms that are more commonly used in regression problems, as demonstrated below:
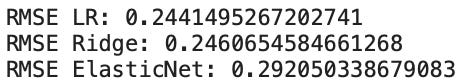
Which produces the following output:
Based on these results we can see that linear regression (LR), ridge regression, and elastic net produce the best results. Thus, these three algorithms can be selected for further evaluation before we decide which one to invest time into tuning and optimizing to enhance our ability to make accurate predictions.
For more on algorithm spot-checking you can check out my previous newsletter titled, Comparing Machine Learning Algorithms.
🧬 Select Evaluation Method and Metric(s):
After spot-checking and selecting the best-performing algorithms, we can perform a more comprehensive evaluation process to decide which algorithm to dedicate more resources towards training.
The three most common evaluation methods are as follows:
Train-test split is a simple evaluation method that splits your dataset into two parts: a training set (used to train your model) and a testing set (used to evaluate your model).
k-fold cross-validation is an evaluation technique that splits your dataset into k-parts to estimate the performance of a machine learning model with greater reliability than a single train-test split; and
Leave one out cross-validation (LOOCV) is a form of k-fold cross-validation, but taken to the extreme where k is equal to the number of samples in your dataset.
If speed is your priority, then a train-test split is often a good choice for evaluating models, so long as the dataset is relatively homogenous. On the other hand, if accuracy and reliability are of the utmost importance, then LOOCV is a good choice as long as your dataset is small to medium-sized (LOOCV is very computationally expensive and thus relatively slow with large datasets).
However, we rarely care only about speed or reliability; in most cases, we want a balance of the two. k-fold cross-validation is helpful for this reason, as it’s both fast and provides accurate results. In fact, k-fold cross-validation (with a k of 3-10) is considered by some to be the gold standard method for evaluating machine learning models due to its versatility and ease of implementation.
For more on evaluation methods you can check out my previous newsletter titled, Evaluating Machine Learning Algorithms.
Next, we need to select an evaluation metric, which is a statistical technique employed to quantify how well the model works. For regression problems, the most common evaluation metrics are as follows:
The mean absolute error (MAE), which is the average of the sum of absolute errors in predicted values;
The root Mean squared error (RMSE), which is the square root of the mean of squared differences between actual and predicted outcomes; and
R², also known as the coefficient of determination, indicates the goodness of fit of a set of predictions compared to the actual values.
In this newsletter, I'm going to use RMSE and R² as my evaluation metrics, which is demonstrated in the code below:
The above code produces the following results:
When using KFCV as an evaluation method and RMSE as an evaluation metric we see that linear regression (LR) performs slightly better than ridge, with elastic net’s performance fairing quite a bit worth than both of the aforementioned algorithms.
The above code produces the following results:
When using KFCV as an evaluation method and R² as an evaluation metric we see that linear regression (LR) performs slightly better than ridge again, with elastic net’s performance significantly falling lagging. As a result, I’ll select linear regression for further tuning and optimization.
For more on evaluation metrics you can check out my previous newsletters titled, Evaluation Metrics For Regression Problems and Evaluation Metrics For Classification Problems.
🧬 Feature Selection:
In a previous article titled Feature Selection For Machine Learning, I used the Oasis-1 dataset to demonstrate the process of selecting features that best predict the model's desired outcome variable. Specifically, I used the following techniques for performing feature selection:
Univariate selection, which is a technique that selects the best data features based on univariate statistical tests;
Recursive feature elimination (RFE), which eliminates the least useful features one by one until a specified number of strong features remain;
Principal component analysis (PCA), which is an unsupervised learning technique used for reducing the dimensionality of a dataset; and
Feature importance, which generates a score for all of the input features, representing their relative importance.
Through that process, I determined that the four most essential features in the dataset were nWBV, MSSE, age, and ASF.
We’ve previously selected linear regression as our algorithm, k-fold cross-validation as our evaluation method, and R² as our evaluation metric. Below you’ll find sample code re-evaluating our model after performing feature selection (i.e., removing all features from our dataset except (nWBV, MSSE, age, and ASF):
Before feature selection, our model produced an R² value of 0.46, whereas, after feature selection, we get an R² of 0.54 (i.e., r value of +0.73), indicating that feature selection improved our model’s performance.
In the next step we’ll transform our dataset, before re-evaluating our model’s performance again.
🧬 Transform Data:
Different machine learning algorithms make different assumptions about input data, and as a result, they require different data pre-processing methods. As a result, it’s essential to know the following before training a machine learning model:
What assumptions your chosen machine learn model makes about your data;
How your data is formatted, how that may differ from the aforementioned assumptions; and
How to pre-process and transform your data so it is formatted appropriately.
Standardization is a data pre-processing technique that centers the distribution of data attributes such that their means and standard deviations are equal. Standardization is a suitable data pre-processing technique when performing linear regression, logistic regression, or linear discriminate analysis.
Below you’ll find sample code standardizing the data in our dataset before re-training and re-evaluating our model:
In many instances, scaling improves a machine learning model’s performance. However, in this specific instance, scaling has little to no effect on the model’s performance. Despite that, scaling is still an important step, making coefficients easier to interpret.
🧬 Tune Hyperparameters:
You can think of machine learning algorithms as systems with various knobs and dials, which you can adjust in any number of ways to change how output data (predictions) are generated from input data. The knobs and dials in these systems can be subdivided into parameters and hyperparameters.
Parameters are model settings that are learned, adjusted, and optimized automatically. Conversely, hyperparameters need to be manually set manually by whoever is programming the machine learning algorithm.
Generally, tuning hyperparameters has known effects on machine learning algorithms. However, it’s not always clear how to best set a hyperparameter to optimize model performance for a specific dataset. As a result, search strategies are often used to find optimal hyperparameter configurations.
If you’re unsure what hyperparemeters you can optimize for a given model you can use the code below (substituting LinearRegression() for your chosen model):
Unfortunately, there are no tunable hyperparameters for linear regression and as a result we will skip this step. However, if you’re interested in learning more about hyperparamter optimization you can check out my previous newsletter titled, An Introduction To Hyperparameter Optimization.
🧬 Create Pipeline To Automate Workflow:
Up to this point, we’ve been working to experimentally derive best practices to improve our machine learning models’ performance. Once sufficiently satisfied with a model’s performance, you can create a pipeline to codify everything you’ve learned and automate the workflow to reproduce your model.
Machine learning pipelines consist of multiple sequential steps that do everything from data extraction and preprocessing to model training and deployment. The following code demonstrates how to create a pipeline that codifies what we’ve learned in our exploration thus far:
As you can see, our model’s performance has also improved from an R² value of 0.54, to 0.58 (i.e., an r value of +0.76). Assuming we’re happy with this models performance we can use it to make predictions on new data. Since we’re using the Oasis-1 dataset, we’ll enter in values for the following features in order to predict whether an individual has diabetes (input values in brackets):
M/F [0]
Age [74]
Edu [3]
SES [3]
MMSE [18]
eTIV [1875]
nWBV [0.65]
ASF [1.3]
The above values are stored in the New_Patient_inputs label and an output is predicted, as demonstrated in the code below:
Remember, a CDR value of 1 means mild cognitive impairment, and a CDR of 2 means moderate cognitive impairment. So, if a new patient presented with the above values and our model predicted a CDR value of 1.2, the patient may suffer from mild cognitive impairment.
In theory, we could use this model to intake a patient's data, as outlined above, and predict their clinical dementia rating (CDR) score. However, in practice, we would want to train our model on a much broader dataset with more individuals of various ages, socioeconomic statuses, ethnicities, and so forth to ensure we're getting a realistic sampling of the population we wish to deploy the model with. Additionally, we would want to include patients with severe cognitive impairment (CDR = 3) in the dataset since our current dataset lacks that (a major limitation in practice). Additionally, we would want to tune our model to increase its performance to avoid type 1 and type 2 errors, which could be highly detrimental when the model is used to make medical and/or lifestyle decisions.
👉 If you have any questions about the content in this newsletter please let me know in the comment section below.
👉 If you liked this post, don’t forget to leave a like ❤️. It helps more people discover this newsletter on Substack. The button is located towards the bottom of this email.
👉 If you enjoy reading this newsletter, please share it with friends!
Paid subscribers can access the Google Collab notebook(s) with all of my code from this article, plus the datasets used, below ⬇️
Keep reading with a 7-day free trial
Subscribe to Decoding Biology by Evan Peikon to keep reading this post and get 7 days of free access to the full post archives.